Determination of phosphoenolpyruvate carboxylase activity
Phosphoenolpyruvate Carboxylase Activity is to understand the function of phosphoenolpyruvate (PEP) shuttle enzyme and to familiarize with the enzyme coupled assay to determine PEP prismase activity.
Principle
The basic principle behind the assay of phosphoenolpyruvate carboxylase activity is that PEP shuttle enzyme is a key enzyme for CO2 fixation in C4 and CAM plants. In the presence of Mg2+, PEP carboxylase catalyzes the formation of oxaloacetic acid (OAA) from PEP and HCO3-, which is catalyzed by malate dehydrogenase (MDH) and can be reduced to malic acid (Mal) by NADH. The reaction was as follows: 
The rate of NADH consumption was calculated by measuring the change in absorbance of the reaction system at 340 nm, and the activity of PEP sauropodase was further deduced.
Operation method
Determination of phosphoenolpyruvate carboxylase activity
Principle
The basic principle behind the assay of phosphoenolpyruvate carboxylase activity is that PEP shuttle enzyme is a key enzyme for CO2 fixation in C4 and CAM plants. In the presence of Mg2+, PEP carboxylase catalyzes the formation of oxaloacetic acid (OAA) from PEP and HCO3-, which is catalyzed by malate dehydrogenase (MDH) and can be reduced to malic acid (Mal) by NADH. The reaction was as follows: the rate of NADH consumption was calculated by measuring the change in absorbance of the reaction system at 340 nm, and the activity of PEP siderophore was further deduced.
Materials and Instruments
Materials: corn, sorghum, etc. C Move The basic procedure for the determination of phosphoenolpyruvate-converting enzyme activity can be divided into the following steps: The leaves were washed and blotted to remove the water, and the midrib was removed. Weigh 20 g, put it in the refrigerator overnight, cut it into pieces the next day and put it into a tissue masher, add 80 mL of extraction buffer (pre-cooled), homogenize for 2 min at 20,000 r/min (running for 30 s, with an interval of 10 s, and repeated homogenization), filter it through 4 layers of gauze, and then take the filtrate into a high-speed freezing centrifuge and centrifuge it at 15,000 g for 10 min, and the supernatant is the crude enzyme extract of PEP sauropodase. The supernatant is the crude enzyme of PEP sauropodase.
4
Leaves of plants.
Reagents:
Extraction buffer: 0.1 mol・L
-1
Tris-H
2
SO
4
buffer (pH 7.4) containing 7 mmol・L
-1
ethanol, 1 mmol・L
-1
EDTA, and 5% glycerol;
Equilibration buffer: 10 mol・L
-1
Tris-H
2
SO
4
buffer (pH 8.2) containing 0.2 mmol・L
-1
EDTA, 0.2 mol・L
-1EDTA, 0.2 mol・L-1
DTT (dithiothreitol), 5% glycerol;
Reaction buffer: 0.1 mol・L
-1
Tris-H
2
SO
4
buffer (pH 9.2) containing 0.1 mol・L
-1
MgCl
2
;
Reaction reagent: 100 mmol・L
-1
NaHCO
3
NaHCO 3, 40 mmol・L-1
-1
PEP, 1 mg・L
-1
NADH (pH 8.9), malate dehydrogenase (MDH).
Equipment:
① UV spectrophotometer
② Freezing centrifuge
③ Tissue masher
④ Sephadex G-25 column (2 cm × 45 cm)
⑤ DEAE (diethylaminoethyl)-cellulose (DE-52, 1 cm × 30 cm) column
⑥ Ultraviolet monitor
⑦ Partial collector
⑦ Peristaltic pump
(1) Step-by-step precipitation of Sulfhydrynium: fill the above crude enzyme solution into a beaker, stir it on a stirrer, slowly add solid Sulfhydrynium powder to reach 35% saturation, let it stand in a refrigerator for 1 h, centrifuge it at 15,000 g for 10 min, then add solid Sulfhydrynium powder into the supernatant to reach 55% saturation, let it stand in a refrigerator for 1 h, and then centrifuge it at 15,000 g for 10 mm, discard the supernatant, and then the precipitate will be recovered. The supernatant was discarded, and the precipitate was reconstituted with 8 mL of equilibrium buffer.
(2) Sephadex G-25 column chromatography: Equilibrate the Sephadex G-25 column (2 cm × 45 cm) with equilibration buffer. The above compounded solution was put on the column, the sample was pressed twice, and eluted with equilibrium buffer at a rate of 50 mL・h-1, and the part with enzyme activity was collected by a detector and centrifuged at 15 000 g for 10 min, and the supernatant was the partially purified enzyme solution of PEP sauropod enzyme.
(3) DEAE-cellulose column chromatography: The transformed DEAE-52 was loaded into a 1 cm × 30 cm chromatography column and equilibrated with equilibration buffer for 2 h. The partially purified enzyme solution mentioned above was put on the DEAE-52 column, pressed the sample for 2 times, and then eluted with equilibration buffer, and then collected after passing through the ultraviolet detector. Then use equilibrium buffer to prepare 0~0.6 mol・L-1 NaCl solution for continuous gradient elution (at a rate of 30 mL・h-1 ), and collect the part with enzyme activity, i.e., the purified PEP invasive enzyme, and use it for enzyme activity determination.
3. Measurement of enzyme activityTake 1 test tube, add 1.0 mL of reaction buffer, 0.1 mL of 40 mmol・L-1 PEP, 0.1 mL of 1 mg・mL-1 NADH (pH 8.9), 0.1 mL of malate dehydrogenase and 0.1 mL of PEP sapogenins (purified extract), and 1.5 mL of distilled water, and then keep warm for 10 minutes in a water bath at a temperature of the measured temperature (e.g. 30 ℃). Then add 0.1 mL of 100 mmol・L-1NaHCO3 to start the reaction, and immediately time the reaction, measure the absorbance value ( A1 ) every 30 s, and record the changes.
4. Experimental results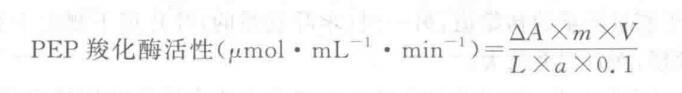
Where: V-Total volume of the assay mixture, 3 mL;
L - optical range of the colorimetric cup, cm.
0.1 a reaction mixture in the amount of enzyme solution, mL;
m - dilution times of enzyme solution;
△A = A0 - A1;
a i molar extinction coefficient of NADH at 340 nm (6.22 × 103 mol-1・cm-1 ).
Caveat
1. The enzyme extraction process should be carried out at 0~4℃.
2. The amount of enzyme solution used in the assay should be tested beforehand. The amount of malate dehydrogenase depends on the amount of PEP sauropodase activity, or the optimal amount can be determined experimentally beforehand.
For more product details, please visit Aladdin Scientific website.
