Determination of ribulose-1,5-bisphosphate carboxylase/oxygenase activity
Ribulose-1,5-bisphosphate synthase/oxygenase (Rubisco) is an important regulatory enzyme of photosynthesis with a dual role. One is to catalyze the fixation of CO2 in the Calvin cycle, and the other is to catalyze the addition of O2 to ribulose-1,5-bisphosphate. Its activity is an important physiological indicator reflecting the growth status, developmental degree and photosynthetic carbon assimilation capacity of plant leaves. The enzyme activity tends to decrease when the plant ages or suffers from adversity. This experiment is to understand its role in photosynthesis and to familiarize with the method of determining Rubi-sco activity.
Principle
The basic principle of the ribulose-1,5-bisphosphate carboxylase/oxygenase activity assay is that ribulose-1,5-bisphosphate carboxylase catalyzes the combination of one molecule of ribulose-1,5-bisphosphate (RuBP) with one molecule of CO2 to produce two molecules of 3-phosphoglycerate (PGA), which can be produced by the action of 3-phosphoglycerate kinase and glyceraldehyde-3-phosphate dehydrogenase. PGA can produce glyceraldehyde-3-phosphate through the action of 3-phosphoglycerate kinase and glyceraldehyde-3-phosphate dehydrogenase and oxidize NADH as follows:
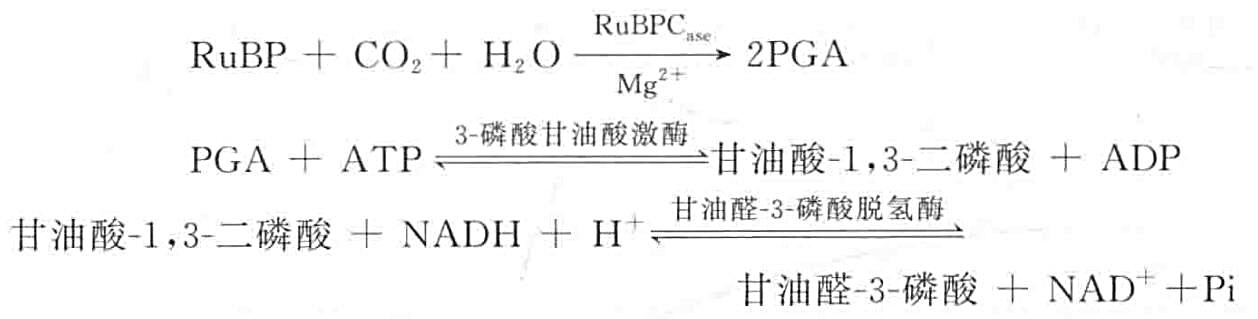
From the above equation, it can be seen that if 1 molecule of CO2 is fixed, 2 molecules of NADH are oxidized. Therefore, the amount of NADH oxidized can be used to calculate the activity of ribulose-1,5-bisphosphate lyase, and the amount of NADH oxidized can be calculated from the change in absorbance at 340 nm. In order to synchronize the oxidation of NADH with CO2 fixation, an ATP regeneration system incorporating creatine phosphate (Cr~P) and phosphocreatine kinase is required.

Ribulose-1,5-bisphosphate oxygenase catalyzes the addition of O2 to the C-2 position of ribulose-1,5-bisphosphate (RuBP) to produce 1 molecule of phosphoglycolic acid and 3-phosphoglyceric acid.The Rubisco oxygenation reaction is a typical monooxygenation reaction in which two oxygen atoms of CO2 are doped into H2O and phosphoglycolic acid, and thus oxygenase activity can be determined by oxygen consumption using the oxygen electrode method. Therefore, oxygenase activity can be determined by oxygen consumption using the oxygen electrode method. In addition, RuBP is first enolated in the oxygenation reaction of the enzyme with the participation of MS2+ ions, at which time the C-2 position of RuBP is adjusted to a negative carbon ion, which when reacted with molecular oxygen forms a peroxide ion, and then electrons return from the oxygen back to the enolated RuBP to generate negative carbon ions again and produce monoclinic oxygen, which can be detected by the luminescence photometer. The luminescence photometer method is 70 times more sensitive than the oxygen electrode method in determining oxygenase activity. However, to determine the absolute value of Rubisco oxygenation activity, the oxygen electrode method must be calibrated first.
Operation method
Determination of ribulose-1,5-bisphosphate carboxylase/oxygenase activity
Principle
The basic principle of ribulose-1,5-bisphosphate carboxylase/oxygenase activity is that ribulose-1,5-bisphosphate carboxylase catalyzes the combination of 1 molecule of ribulose-1,5-bisphosphate (RuBP) with 1 molecule of CO2 to produce 2 molecules of glycerol-glycolinate 3-phosphate (PGA), which is oxidized to produce glyceraldehyde-3-phosphate (NADH) by the action of glycerol-glycolinate-3-phosphate kinase (GGK) and glyceraldehyde-3-phosphate dehydrogenase (GADH). PGA can produce glyceraldehyde-3-phosphate through the action of 3-phosphoglycerate kinase and glyceraldehyde-3-phosphate dehydrogenase, and oxidize NADH as follows: From the above equation, it can be seen that if 1 molecule of CO2 is fixed, 2 molecules of NADH are oxidized. Therefore, the activity of ribulose-1,5-bisphosphate clonase can be calculated from the amount of NADH oxidized, and the amount of NADH oxidized can be calculated from the change in absorbance at 340 nm. In order to synchronize NADH oxidation with CO2 fixation, an ATP regeneration system incorporating creatine phosphate (Cr~P) and phosphocreatine kinase is required. Ribulose-1,5-bisphosphate oxygenase catalyzes the addition of O2 to the C-2 position of ribulose-1,5-bisphosphate (RuBP) to produce 1 molecule of phosphoglycolic acid and 3-phosphoglyceric acid.The Rubisco oxygenation reaction is a typical monooxygenation reaction in which two oxygen atoms of CO2 are doped into H2O and phosphoglycolic acid, and thus oxygenase activity can be determined by oxygen consumption using the oxygen electrode method. Thus, oxygenase activity can be determined by oxygen consumption using the oxygen electrode method. In addition, RuBP is first enolated in the oxygenation reaction of the enzyme with the participation of MS2+ ions, and then the C-2 position of RuBP is adjusted to a negative carbon ion, which reacts with molecular oxygen to form a peroxide ion, and then the electrons return from the oxygen to the enolated RuBP to generate negative carbon ions again and produce a single line of oxygen, which can be detected by the luminescence photometer. The luminescence photometer method is 70 times more sensitive than the oxygen electrode method in determining oxygenase activity. However, to determine the absolute value of Rubisco oxygenation activity, the oxygen electrode method must be calibrated first.
Materials and Instruments
Material: leaves of spinach, wheat and rice plants. Move The basic procedure for the determination of ribulose-1,5-bisphosphate carboxylase/oxygenase activity can be divided into the following steps: 1. Preparation of enzyme crude extract: take 10 g of fresh spinach leaves, wash and dry them, put them into a homogenizer, add 10 mL of pre-cooled extraction medium, homogenize at high speed for 30 s, stop for 30 s, and do it alternately for 3 times; homogenize is filtered through 4 layers of gauze, and the filtrate is centrifuged at 20,000 g for 15 min under 4 ℃, and the precipitate is discarded; the supernatant is the enzyme crude extract and is preserved at 0 ℃ for spare use. 2. Determination of RuBPCase activity: Prepare the enzyme reaction system according to Table 15-1. Table 15-1: Solvent content and preparation. Reagent Amount Reagent Additive amount 5 mmol・ml-1 NADH 0.2 Reaction medium 1.4 50 mmol・ml-1 ATP 0.3 160 U・ml-1 Phosphocreatine kinase 0.1 Extraction medium 0.1 160 U・ml-1 phosphoglycerate kinase 0.1 50 mmol・ml-1 phosphocreatine 0.2 160 U・ml-1 Glyceraldehyde-3-phosphate dehydrogenase 0.2 160 U・ml-1 Glyceraldehyde-3-phosphate dehydrogenase 0.2 mol・ml-1NaHCO3 0.2 Distilled water 0.3 3. Shake the prepared reaction system well and pour it into a colorimetric cup, use distilled water as blank, and take the absorbance of the reaction system at 340 nm on the UV spectrophotometer as the zero point value. The absorbance of the reaction system at 340 nm on the UV spectrophotometer was used as the zero point value. 0.1 mL of RuBP was added into the colorimetric cup and the absorbance was measured every 30 s for 3 min. The enzyme activity was calculated by the absolute value of the decrease in absorbance from the zero point to the first minute.
Reagent:
5 mmol・L
-1
NADH; 25 mmol・L
RuBP; 0.2 mol・L-1
RuBP; 0.2 mol・L-1 NADH
-1
NaHCO
3
Extraction medium: 40 mmol・L
Extraction medium: 40 mmol・L
-1
Tris-HCl buffer (pH 7.6) containing 10 mmol・L -1
-1
MgCl
2
0.25 mmol・L
-1
EDTA, 5 mmol・L
-1
glutathione.
Reaction medium: 100 mmol・L
-1
Tris-HCl buffer (pH 7.8) containing 12 mmol・L -1
-1
MgCl
2
and 0.4 mmol-L-1 Tris-HCl buffer (pH 7.8) containing 12 mmol-L-1 MgCl2
-1
EDTA-Na
2
and 0.4 mmol・L-1 EDTA-Na 2; 160 U・mL
-1
Phosphocreatine kinase solution; 160 U・mL-1
-1
Oleoyl-3-acid dehydrogenase solution; 50 mmol・L
-1
ATP; 50 mmol・L -1
-1
Creatine phosphate; 160 U・mL
-1
Phosphoglycerate kinase solution.
Extraction buffer: 100 mmol・L
-1
Tris-HCl (pH 7.8); 1 mmol・L
-1
EDTA; 20 mmol・L
-KC1.
KC1.
Resuspension buffer: 25 mmol・L
-1
Tris-HCl (pH 7.8); 1 mmol・L-1
-1
EDTA; 5 mmol・L
-1
ethanol; 20 mmol・L-1
-1
KCl.
Oxygen electrode method reaction solution: 100 mmol・L-1
-1
Tris-HCl (pH 8.2); 0.4 mmol・L
-1
EDTA; 20 mmol・L
-1
MgCl
2
Luminometer reaction solution: 50 mmol・L-1 MgCl2
Luminescence photometry reaction solution: 50 mmol・L-1
-1
Tris-HCl (pH 8.0); 1.0 mmol・L -1
-1
MnCl
2
MnCl2; 1.0 mmol・L
-1
NaHCO
3
.
Sodium sulfite; 0.1 mmol・L
-1
Dithiothreitol (DTT); Hyun Sulfate; NaCI; 10 mmol・L
-1
RuBP stock solution (pH 6.5).
Equipment:
① UV spectrophotometer
② Freezing centrifuge
③ Tissue masher
④ Pipette
⑤ Stopwatch
⑥ Oxygen electrode oxygen measurement device
⑦ FG-300 Luminescence Photometer
Since PGA may be present in the enzyme extraction medium, which may cause errors in the determination of enzyme activity, it is necessary to make a sample without RuBP in addition to the above determination.
In addition to the above assay, a control without RuBP was made. The reaction system of the control is exactly the same as that of the above enzyme reaction system, except that the enzyme extraction medium is added at the end, and the absorbance at 340 nm is measured immediately after the addition of the enzyme extraction medium, and the change in absorbance during the first minute is recorded, which should be subtracted from the calculation of enzyme activity.
4. Calculation of results

Where: ∆A - the absolute value of the change in absorbance at 340 nm during the first 1 min of the reaction (minus the change in the control solution during the first 1 min);
N - number of dilutions;
6.22 - absorbance coefficient at 340 nm per micromole of NADH;
2-indicates that 2 mol of NADH is oxidized for every 1 mol of CO2 fixed;
∆t-measurement time 1 min;
d - optical range of the cuvette, cm.
(II) Determination of oxygenation activity of ribulose-1,5-bisphosphate synthase/oxygenase1. Extraction and purification of Rubisco: Take 5~7 g of leaf blade and add 10 mL of pre-cooled extraction buffer to 4 ℃, homogenize, centrifuge at 15 000 g for 10 min, and take the supernatant for spare.
The crude extract obtained above was precipitated step by step with 40% saturated sulfuric acid hyun, and frozen centrifuged for 20 min (4 ℃, 8 000 g). Then the supernatant was taken and 70% saturated Sulfuric acid was added, and the precipitate was taken after freezing and centrifugation for 20 min (4 ℃, 10 000 g), and dissolved with a small amount of resuspension buffer, and then desalted by Sephadex G-25 on a DEAE (DE 52) column, and then the precipitates were collected step by step by gradient elution with resuspension buffer containing 0.20~0.5 mmol・ml-1 NaCl, and then collected by step by gradient elution with 0.20~0.25 mmol・ml-1 NaCl. 0.25 mmol・ml-1 NaCl. 70% sulfuric acid precipitate was stored at 20 ℃, and then dissolved in resuspension buffer at 8 mg・ml-1 before the activity was measured.
2. Oxygen electrode assay for the determination of Rubisco oxygenase activity: 2 mL of the reaction solution of the oxygen electrode assay was stirred in the atmosphere for 10 min, so as to equilibrate the dissolved oxygen in the solution with the atmosphere. Then put the electrode on the reaction chamber, adjust the sensitivity knob of the oxygen meter, so that the recorder to the full scale, and then add 0.1 mL of saturated sodium sulfite, in addition to the oxygen in the water, the pointer returned to close to 0, according to the pointer back to the number of frames and the amount of dissolved oxygen in the water at 25 ℃, it can be calculated for each frame of the record of the amount of oxygen dissolved in water paper represents. The amount of dissolved oxygen in water at 25 °C is 0.26 μmol・mL-1. The amount of dissolved oxygen represented by each grid is calculated as follows:

Add 2 mL of air-saturated solution to the reaction chamber, add 0.1 mL of enzyme solution (8 mg・ml-1 ), hold the solution at 25 ℃ for 10 min, install the electrode, and record the blanking rate of the oxygen consumed by DTT oxidation, and then add 0.01 mL of RuBP stock solution to start the reaction, and then record the reaction speed, which is expressed in terms of the number of squares of recording paper per minute. The activity of oxygenase can then be calculated by the following formula:
3. Determination of Rubisco oxygenase activity by luminescence photometer method: first add 25 μL of RuBP stock solution into the colorimetric cup, put it into the dark room of luminescence photometer reaction, then put 1.4 mL of luminescence photometer reaction solution with 10 μL of the enzyme solution in the mixture at 25 ℃ and keep warm for 10 min, and then inject it into the colorimetric cup to start the reaction, and then record the luminescence curve automatically. The luminescence intensity was expressed as peak height (mm).
Caveat
1. Extraction of Rubisco should be carried out in an ice bath.
2. RuBP is very unstable, especially in alkaline environments, so it should be stored at - 20 °C between pH 5.0-6.
5, preferably ready to use.
For more product details, please visit Aladdin Scientific website.

