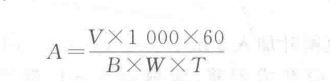Determination of superoxide dismutase (SOD) activity
To learn and understand the method and principles of the photochemical reduction method of nitro nitro tetrazolium blue chloride (NBT) for the determination of SOD activity.... And understand the action characteristics of SOD.
Principle
1. The production of free radicals is favored by the disruption of the metabolic balance of free radicals in cells during adversity stress or senescence in plants. Free radicals are atoms or groups of atoms with unpaired valence electrons. The free radicals produced in living organisms are mainly superoxide radicals ( O2-・ ), hydroxyl radicals (OH・), peroxyl radicals (ROD), and alkoxyl radicals (RO).
Plant cell membranes have two types of peroxide defense systems, enzymatic and non-enzymatic, and superoxide dismutase (SOD), catalase (CAT), peroxidase (POD) and ascorbate peroxidase (ASA-POD) are important protective enzymes of the enzymatic defense system. Ascorbic acid (vitamin C), vitamin E and reduced glutathione (GSH) are important antioxidants in the non-enzymatic defense system. The levels of reactive oxygen scavengers, such as CAT, and reactive oxygen species, such as O2-, H2O2, OH- and O2, can be used as physiological and biochemical indicators of plant senescence.
Superoxide dismutase (SOD) is a metal cofactor-containing enzyme. Higher plants contain two types of SoD: Mn-SOD and Cu. Zn-SOD, which can scavenge superoxide radicals ( O2-・ ) from biological cells through the disproportionation reaction to produce H2O2 and O2, which is catalyzed by catalysis of catalase (CAT) to produce H2O and O2. This reduces the toxic effects of free radicals on organisms. The reaction formula is as follows:
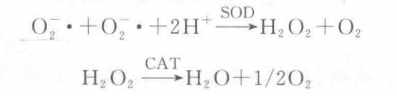
Since superoxide radicals (O broad) are unstable radicals with extremely short lifetimes, the determination of SOD activity is generally an indirect method and utilizes a variety of color-presenting reactions to determine the viability of SOD. Riboflavin can produce superoxide radical negative ion O2-・ under aerobic conditions, and when added to NBT, under light conditions, it reacts with superoxide radical to produce monomethyl meal (yellow color), and then reduces to produce dimethyl meal, which is a blue color substance with maximum absorption peak at 560 nm wavelength.
When SOD was added, it could make the superoxide radical combine with H+ to generate H2O2 and O2, thus inhibiting the photoreduction of NBT and slowing down the generation of blue dimethyl wax. The relative percentage of inhibition of NBT photoreduction was proportional to the enzyme activity within a certain range by adding different amounts of SOD enzyme solution into the reaction solution, and the relative percentage of inhibition of NBT photoreduction was plotted on the coordinate paper with the amount of enzyme solution as the horizontal coordinate and the relative percentage of inhibition of NBT photoreduction as the vertical coordinate, and the correlation curves were plotted on the coordinate paper, and the enzyme activity was calculated based on the relative percentage of inhibition of NBT photoreduction by SOD. The relative percentage of NBT photoreduction inhibition by SOD was used to calculate the enzyme activity. The enzyme activity was calculated according to the relative percentage of NBT photoreduction inhibition by SOD. The amount of enzyme when the relative percentage of NBT photoreduction inhibition by SOD was 50% was found to be a unit of enzyme activity (U).
Operation method
Determination of superoxide dismutase (SOD) activity
Principle
1. The production of free radicals is favored by the disruption of the metabolic balance of free radicals in cells during adversity stress or senescence in plants. Free radicals are atoms or groups of atoms with unpaired valence electrons. The free radicals produced in living organisms are mainly superoxide radicals (O2-・), hydroxyl radicals (OH・), peroxyl radicals (ROD), and alkoxyl radicals (RO). Plant cell membranes have two types of peroxide defense systems, enzymatic and non-enzymatic, and superoxide dismutase (SOD), catalase (CAT), peroxidase (POD) and ascorbate peroxidase (ASA-POD) are important protective enzymes of the enzymatic defense system. Ascorbic acid (vitamin C), vitamin E and reduced glutathione (GSH) are important antioxidants in the non-enzymatic defense system. The levels of reactive oxygen scavengers, such as CAT, and reactive oxygen species, such as O2-, H2O2, OH- and O2, can be used as physiological and biochemical indicators of plant senescence. Superoxide dismutase (SOD) is a metal cofactor-containing enzyme. Higher plants contain two types of SoD: Mn-SOD and Cu. Zn-SOD, which can scavenge superoxide radicals (O2-・) from biological cells through a disproportionation reaction to produce H2O2 and O2, which is catalyzed by catalysis of catalase (CAT) to produce H2O and O2. This reduces the toxic effects of the free radicals on the organism. The reaction formula is as follows: Since the superoxide radical (O wide) is an unstable radical with a very short life span, the determination of SOD activity is generally an indirect method and utilizes a variety of color-presenting reactions to determine the viability of SOD. Riboflavin can produce superoxide radical negative ion O2-・ under aerobic condition, when added to NBT, under the light condition, it reacts with superoxide radical to produce monomethyl meal (yellow color), and then reduces to produce dimethyl wax, which is a blue color substance with maximum absorption peak at 560 nm wavelength. When SOD was added, it could make the superoxide radical combine with H+ to generate H2O2 and O2, thus inhibiting the photoreduction of NBT and slowing down the generation of blue dimethyl wax. The relative percentage of inhibition of NBT photoreduction was proportional to the enzyme activity within a certain range by adding different amounts of SOD enzyme solution into the reaction solution, and the relative percentage of inhibition of NBT photoreduction was plotted on the coordinate paper with the amount of enzyme solution as the horizontal coordinate and the relative percentage of inhibition of NBT photoreduction as the vertical coordinate, and the correlation curves were plotted on the coordinate paper, and the enzyme activity was calculated based on the relative percentage of inhibition of NBT photoreduction by SOD. The relative percentage of NBT photoreduction inhibition by SOD was used to calculate the enzyme activity. The enzyme activity was calculated according to the relative percentage of NBT photoreduction inhibition by SOD. The amount of enzyme when the relative percentage of NBT photoreduction inhibition by SOD was 50% was found to be a unit of enzyme activity (U).
Materials and Instruments
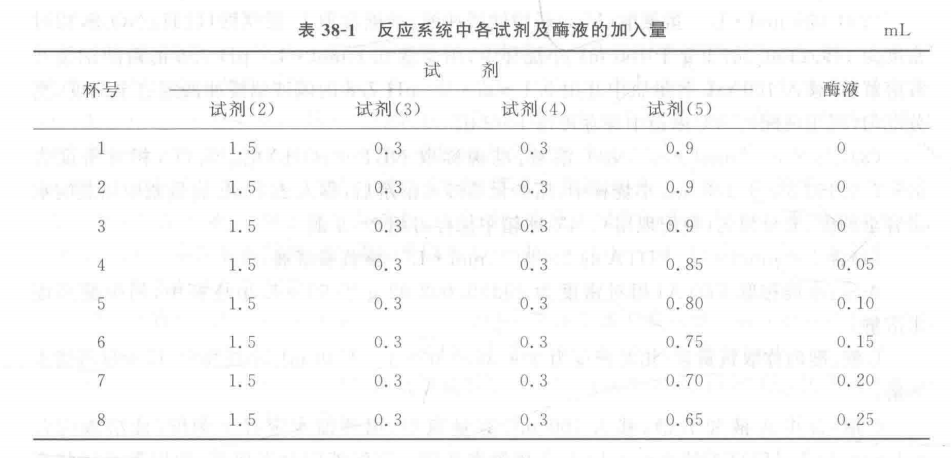
1. Material: Fresh leaves of wheat, corn, rice and cotton. Move The basic procedure for the determination of superoxide dismutase (SOD) activity can be divided into the following steps. 1: 1. Preparation of enzyme solution: Add 3 mL of 0.05 mol - L-1pH 7.8 sodium phosphate buffer per gram of fresh leaves, add a small amount of quartz sand, and grind it into a homogenate in a mortar and pestle in an ice bath, condense it into a 5 mL graduated centrifuge tube, and then centrifuge it for 30 min at 8,500 r - min-1 (10,000 g), and the supernatant will be the crude extract of SOD enzyme. 2. Determination of enzyme activity: For each treatment, take 8 washed and dried micro beakers, add each reagent and enzyme solution according to Table 38-1, and the total volume of the reaction system is 3 mL, in which the amount of sodium phosphate buffer and enzyme solution in No. 4-8 cups can be adjusted according to the concentration of the enzyme solution in the test material and the enzyme activity (e.g., the enzyme concentration is large and the enzyme activity is strong, the enzyme dosage will be reduced appropriately). After all the reagents are added, mix thoroughly, take the No. 1 micro-beaker and put it in the dark, as a blank control, and use it for zero adjustment during colorimetry. The remaining 7 microbeakers were placed in a light box (installed with 3 fluorescent tubes of 20 W) at a temperature of 25 ℃ and a light intensity of 4 500 lx for 15 min, and then the reaction was terminated by blocking the light immediately. The reaction was terminated by shading immediately. The optical density of each cup was measured at 560 nm by zero adjustment with cup No. 1 and the results were recorded. The average of the optical densities of No. 2 and No. 3 cups was taken as the 100% inhibition of NBT photoreduction, and the relative percentages of inhibition of NBT photoreduction were calculated from the optical densities of the other cups with different amounts of enzyme solution in each reaction system. Take the amount of enzyme solution as the horizontal coordinate and the relative percentage of inhibition of NBT photoreduction as the vertical coordinate, and make the correlation curve between the two. The amount of enzyme solution with 50% inhibition (ptL) was found as a unit of enzyme activity (U). 1. 1. Calculation of results: (1) Measure the optical density of each cup of liquid at 560 nm and fill in Table 38-2. Take the amount of enzyme solution added as the horizontal coordinate and the relative percentage of inhibition of NBT photoreduction as the vertical coordinate, and plot the correlation curve between the two on the coordinate paper. Find the amount of enzyme solution with 50% inhibition (卩L) as a unit of enzyme activity (U). (2) SOD enzyme activity is calculated according to the following formula: V - total volume of enzyme extract, mL; B - amount of enzyme solution for one enzyme vigor unit, μL; W a sample fresh weight, g; T a reaction time, min; 1 000-1 mL=1000 μL 60 a 1 h=60 min . The inhibition rate was calculated according to the following formula: Where: 9- Mean value of optical density of cups 2 and 3 liquid; D?i optical density value of each cup liquid with different enzyme liquid amount added. Note: Sometimes due to the large number of samples, each sample according to this method to determine the enzyme activity workload will be very large, can also be determined for each sample only 1 or 2 enzyme dosage of optical density values, according to the following formula to calculate the enzyme activity. Where: D1 a 2, 3 cup of liquid optical density of the average value; D2 - the optical density of the sample enzyme solution; 50% - inhibition rate of 50% ; Other factors represent the same content as the factors represented in the above SOD enzyme activity calculation formula. Caveat 1. Phenolic-rich plants (such as tea) in the homogenization of a large number of polyphenols, will cause irreversible precipitation of the enzyme protein, so that the enzyme inactivation, so in the extraction of such plants SOD enzyme, must be added to the adsorbent polyphenols, polyphenols to remove, to avoid denaturation and inactivation of enzyme proteins, generally in the extraction solution with 1% ~ 4% of the polyvinylpyrrolidone (PVP). 2. The temperature and photochemical reaction time must be strictly controlled. In order to ensure that the light intensity of each micro beaker is consistent, all micro beakers should be arranged in a straight line parallel to the fluorescent tube. For more product details, please visit Aladdin Scientific website.
2. Reagents.
(1) 0.1 mol ・ L
-1
pH 7.8 sodium phosphate (N
2
HPO
4
-NaH
2
PO
4
) buffer:
Solution A (0.1 mol - L
-1
Na
2
HPO
4
solution): accurately weigh the Na
2
HPO
4
・12H
2
O (relative density of 358.14) 3.5814 g in a 100 mL beaker, dissolved with distilled water, transferred to a 100 mL volumetric flask and fixed to the scale with distilled water, mixed thoroughly. 4 ° C refrigerator to keep for spare.
Liquid B (0.1 mol・L
-1
NaH
2
PO
4
solution): accurately weigh the NaH
2
PO
4
・2H
2H
O (relative density 156.01) 0.780 g in a 50 mL beaker, dissolved with a small amount of distilled water, transferred to a 50 mL volumetric flask with distilled water to the moment, mixed thoroughly. 4 ° C refrigerator to keep spare.
Take 183 mL of the above liquid A and 17 mL of liquid B and mix well to form 0.1 mol - L
-1
Mix 183 mL of solution A with 17 mL of solution B to make 0.1 mol - L -1 sodium phosphate buffer pH 7.8. Store in a refrigerator at 4°C.
(2) 0.026 mol - L
-1
Methionine (Met) sodium phosphate buffer: accurately weigh the L-methionine (C
5
H
11
NO
S, relative density 149.21) 0.3879 g in a 100 mL beaker.
S,relative density 149.21) 0.3879 g in a 100 mL beaker with a small amount of 0.1 mol - L
-L
After dissolved with a small amount of 0.1 mol - L -1 sodium phosphate buffer pH 7.8, transferred to a 100 mL volumetric flask and dissolved with 0.1 mol - L -1 sodium phosphate buffer pH 7.8.
L-1
pH 7.8 sodium phosphate buffer to the scale, mix well (ready to use). 4 ° C refrigerator storage can be used for 1 ~ 2d.
(3) 7.5×10-4
-4 mol・L
mol・L
-1) NBT solution: accurately weigh the NBT solution.
(3) 7.5×10 -4 mol・L -1 NBT solution: accurately weigh the NBT (C
4
OH
3
OCl
OCl
N
10
O
6
(relative density 817.7) 0.153 3 g in a 100 mL beaker, dissolved with a small amount of distillation, transferred to a 250 mL volumetric flask with distilled water to the scale, mix thoroughly (ready to use). 4 ° C refrigerator storage can be used for 2-3 d. (4) Containing 1 0 μmol ・ L of the solution.
(4) Containing 1.0 μmol ・ L
-1.0 μmol ・ L
L-1 of EDTA.
(4) 2×10×L-1 EDTA containing 1.0 μmol ・ L -1 -5
L L
-1
Riboflavin solution:
Solution A: Accurately weigh 0. 00292 g of EDTAC (relative density of 292) in a small 50 mL beaker and dissolve with a small amount of distilled water.
Liquid B: accurately weigh riboflavin (relative density of 376.36) 0.0753 g in a small beaker of 50 mL, with a small amount of distilled water to dissolve.
Liquid C: Combine liquid A and liquid B, transfer to a 100 mL volumetric flask, and then dilute with distilled water to the scale.
-1
This solution is a 2 mmol-L-1 solution containing 0.1 mmol-L-1 EDTA.
-1
Riboflavin solution containing 0.1 mmol - L -1 EDTA. The solution should be kept away from light, i.e., the brown bottle containing the solution should be wrapped with black paper and kept in the refrigerator at 4°C for 8-10 d. The solution should be kept in the refrigerator at 4°C for 8-10 d.
When determining the SOD enzyme activity, the solution C was diluted 100-fold, i.e., the solution contained 1.0 μnmol - L -1
-L
L-1 EDTA.
-5
L-1EDTA.
-1
riboflavin solution.
(5) 0.05 mol - L
-1
pH 7.8 sodium phosphate buffer: take 0.1 mol - L
-1
pH 7.8 sodium phosphate buffer 50 mL, transferred to a 100 mL volumetric flask with distilled water to the gradient, mix thoroughly. 4 ° C refrigerator to save standby.
(6) Quartz sand.
3. Equipment: spectrophotometer, analytical balance, high-speed freezing centrifuge, refrigerator, 4500 lx light box, ceramic plate with lid, pipette rack, mortar, 5 mL centrifugal tube, 10-15 mL micro beaker, 0.5 mL, 1 mL, 2 mL, 5 mL pipette or sampling device, 50 μL, 100 μL microsampler, 50 mL, 100 mL, 500 mL, 1,000 mL beaker, 50 mL, 500 mL, 1,000 mL beaker. 50 mL, 100 mL, 500 mL, 1,000 mL beakers, 50 mL, 100 mL measuring cylinders, 50 mL, 100 mL, 250 mL, 1000 mL volumetric flasks, 125 mL spouted bottles.